Malware Classification
Using a CNN to classify MalwareImages
Deep Learning Project - Reproduction Study
In this assignment, we reproduced the CNN presented in “Using convolutional neural networks for classification of malware represented as images”* [1]. This CNN was then tested using 10-fold cross validation to determine whether we would achieve similar performance to the paper. Furthermore, we implemented nested cross validation to perform further hyperparameter tuning on the CNN. The best model architecture that was found was then trained and tested on a much larger dataset (~147GB) of malware images.

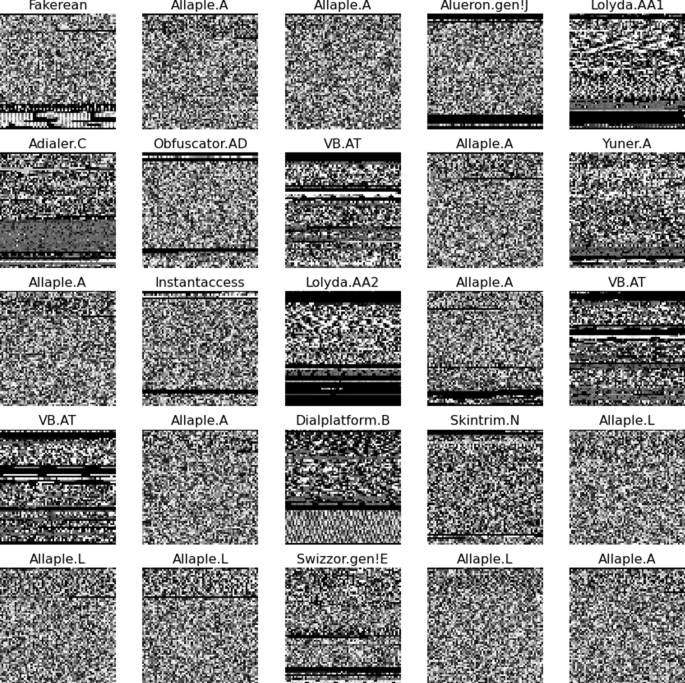
Hyperparameter Tuning
Nested cross-validation was used in grid search to mitigate overfitting and yield a more dependable assessment of model performance. Regular cross-validation may lead to higher estimates of model performance because the model evaluation is done on seen data. Nested cross-validation performs an outer loop of cross-validation for model evaluation and an inner loop of cross-validation for hyperparameter tuning, ensuring that the model’s performance is evaluated on unseen data during hyperparameter tuning. This helps to provide a more realistic estimation of the model’s performance.
| Number of Convolutional Layers | Number of Feed Forward Layers | Number of Output Nodes | FeedForward Layer Output Sizes | Kernel Size for Convolution Layer |
|---|---|---|---|---|
| 2 | [1, 2] | [40, 50], [70, 90] | [25], [50, 25] | [5, 3], [9, 5] |
| 3 | [1, 2] | [25, 50, 50], [50, 70, 70], [70, 90, 90] | [25], [50, 25] | [5, 3, 3], [9, 5, 5] |
| 4 | [1, 2] | [20, 30, 40, 50], [50, 70, 70, 90] | [25], [50, 25] | [5, 3, 3, 3], [9, 5, 5, 3] |
Running the code
After cloning the respository, you should have the following files:
-
CNN.pyThis file contains the CNN class. You should not have to change anything in this file. -
nested_cross_validation.pyThis file contains the logic for the nested cross validation. You should not have to change anything in this file. -
cross_validation.pyThis file contains the logic for the cross validation. You should not have to change anything in this file. -
parameters.pyThis file contains the parameters and transforms for the models that were used to get the results. You should not have to change anything in this file. -
config.pyThis file contains some paths to the datasets and in this file you also set some parameters that are used bynested_crosss_validation.pyandcross_validation.py. You will have to change this file.
Changes that you need to make in config.py
MALIMG_DATASET_PATH and AKASH_DATASET_PATH store the paths to the MalImg dataset and Akash’s dataset, respectively. Hence, you need to set these paths correctly, such that the datsets can be accessed by the code.
CV_DATASET_PATH specifies the path to the dataset that you want to run. CV_DATASET_PATH should be set equal to either MALIMG_DATASET_PATH or AKASH_DATASET_PATH. Later will be explained how this variable should be assigned.
CV_PARAMS specifies the model that you want to run. CV_PARAMS should be set equal to either base_model_malimg, best_model_malimg, or best_model_dataAkash. Note that if you selected the MalImg dataset in CV_DATASET_PATH, you can only use the models base_model_malimg and best_model_malimg. Similarly, if you specified Akash’s dataset to be used, you can only use the model best_model_dataAkash. These model parameters are stored inside parameters.py. base_model_malimg contains the parameters of the CNN from the paper. best_model_malimg contains the parameters of the best CNN that we found in the nested_cross validation. best_model_dataAkash contains the parameters of the best CNN that we found in the nested_cross validation, but this model is meant to be run only by cross_validation.py on Akash’s dataset.
The variables below specify the paths to store the models that are stored and loaded in nested_cross_validation.py. Please set these paths appropriately.
INNER_MODEL_PARAMS_PATH = 'inner_model_params.pt'
INNER_MODEL_PATH = 'working/best_inner_model.pt'
OUTER_MODEL_PATH = 'best_outer_model.pt'
OUTER_MODEL_PARAMS_PATH = 'outer_model_params.pt'
LOG_FILE = 'log_file.txt'
Our runs
To get the results that are shown in the Results section, the following runs were executed:
- Cross validation on
base_model_malimgon the MalImg dataset
To reproduce the results of the paper, we ran cross_validation.py on the MalImg dataset with the baste_model_malimg parameters. To replicate this run, set the following parameters in the config.py file as follows:
CV_DATASET_PATH = MALIMG_DATASET_PATH
CV_PARAMS = base_model_malimg
All other parameters are already set to the same parameters that were used to run it by use. However, if you need to change the batch size, you can change batch_size inside cross_validation.py. Once the config.py file is set correctly, you simply need to run cross_validation.py.
- Nested cross validation on the MalImg dataset
Next, we performed hyperparameter tuning to try and find a better model on the MalImg dataset. You can replicate this run by simply running nested_cross_validation.py. This will automatically run the grid search with the parameter grid stored inside parameters.py on the MalImg dataset, with the same batch size, number of epochs, number of inner folds, and number outer folds that we ran it at. You can change the batch size by changing the variable BATCH_SIZE inside nested_cross_validation.py. Note that CV_DATASET_PATH and CV_PARAMS are not used by nested_cross_validation.py, since this code should only be run on the MalImg dataset. Hence, you do not need to change any variables inside config.py for this run.
- Cross validation with best model on MalImg
The parameters of the best model resulting from the nested cross validation is stored in best_model_malimg. Hence, to replicate this run, set the variables in config.py as follows:
CV_DATASET_PATH = MALIMG_DATASET_PATH
CV_PARAMS = best_model_malimg
And then run cross_validation.py.
- Cross validation with best model on Akash’s dataset
Finally, the best model resulting from the hyperparameter tuning was trained and tested on Akash’s dataset. To replicate this run, set the config.py file as follows:
CV_DATASET_PATH = AKASH_DATASET_PATH
CV_PARAMS = best_model_dataAkash
and run the cross_validation.py file.
Results
Grid Search on Malimg Dataset
From the grid search a few observations were made. Below are the best models found during the nested cross validation. As can be seen in the table, the architecture with 3 convolutional layers performed the best. However, the architecture with only 2 convolutional layers was not far off in terms of test accuracy which was intriguing to see. Another thing that was noted is how adding 2 feed forward layers greatly decreased the performance of the model.
Best Parameters
| Convolutional Layers | Feed Forward Layers | Output Nodes | Output Sizes | Kernel Size |
|---|---|---|---|---|
| 4 | 1 | [50, 70, 70, 90] | [25] | [5, 3, 3, 3] |
| 3 | 1 | [70, 90, 90] | [25] | [5, 3, 3] |
| 2 | 1 | [40, 50] | [25] | [5, 3] |
Accuracy, MAE and Std. Dev for Convolutional Layers
| Number of Convolutional Layers | Test Accuracy | Test MAE | Std. Dev. Accuracy | Std. Dev. MAE |
|---|---|---|---|---|
| 4 | 97.27% | 0.30 | 0.151 | 0.017 |
| 3 | 98.62% | 0.07 | 0.149 | 0.004 |
| 2 | 98.52% | 0.07 | 0.084 | 0.009 |
Hence, the best model resulting from the hyperparameter tuning with nested cross validation was the CNN with 3 convolutional layers, output channels [70,90,90], kernel sizes [5,3,3], and a single feedforward layer. Once we had the best model it was ran on the Malimg dataset in order to compare the architecture that the paper proposes against the architecture that was found through grid search.
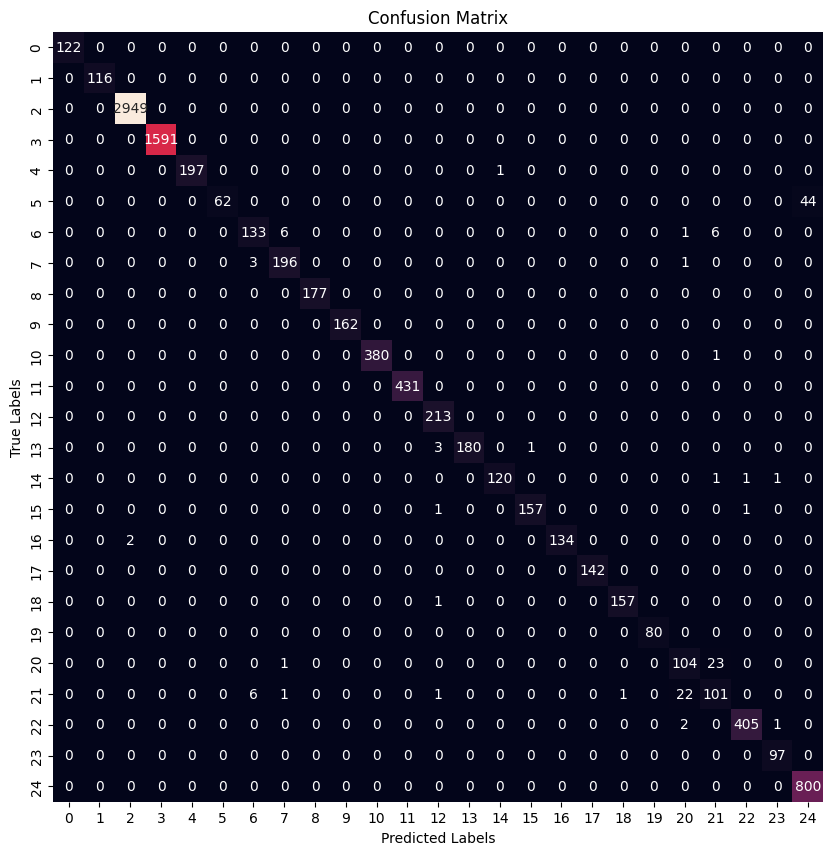
Paper’s Model Results
| Precision | Recall | F1 Score | |
|---|---|---|---|
| 0 | 1.000000 | 1.000000 | 1.000000 |
| 1 | 1.000000 | 1.000000 | 1.000000 |
| 2 | 0.999322 | 1.000000 | 0.999661 |
| 3 | 1.000000 | 1.000000 | 1.000000 |
| 4 | 1.000000 | 0.994949 | 0.997468 |
| 5 | 1.000000 | 0.584906 | 0.738095 |
| 6 | 0.936620 | 0.910959 | 0.923611 |
| 7 | 0.960784 | 0.980000 | 0.970297 |
| 8 | 1.000000 | 1.000000 | 1.000000 |
| 9 | 1.000000 | 1.000000 | 1.000000 |
| 10 | 1.000000 | 0.997375 | 0.998686 |
| 11 | 1.000000 | 1.000000 | 1.000000 |
| 12 | 0.972603 | 1.000000 | 0.986111 |
| 13 | 1.000000 | 0.978261 | 0.989011 |
| 14 | 0.991736 | 0.975610 | 0.983607 |
| 15 | 0.993671 | 0.987421 | 0.990536 |
| 16 | 1.000000 | 0.985294 | 0.992593 |
| 17 | 1.000000 | 1.000000 | 1.000000 |
| 18 | 0.993671 | 0.993671 | 0.993671 |
| 19 | 1.000000 | 1.000000 | 1.000000 |
| 20 | 0.800000 | 0.812500 | 0.806202 |
| 21 | 0.765152 | 0.765152 | 0.765152 |
| 22 | 0.995086 | 0.992647 | 0.993865 |
| 23 | 0.979798 | 1.000000 | 0.989796 |
| 24 | 0.947867 | 1.000000 | 0.973236 |
Best Model Found From Grid Search
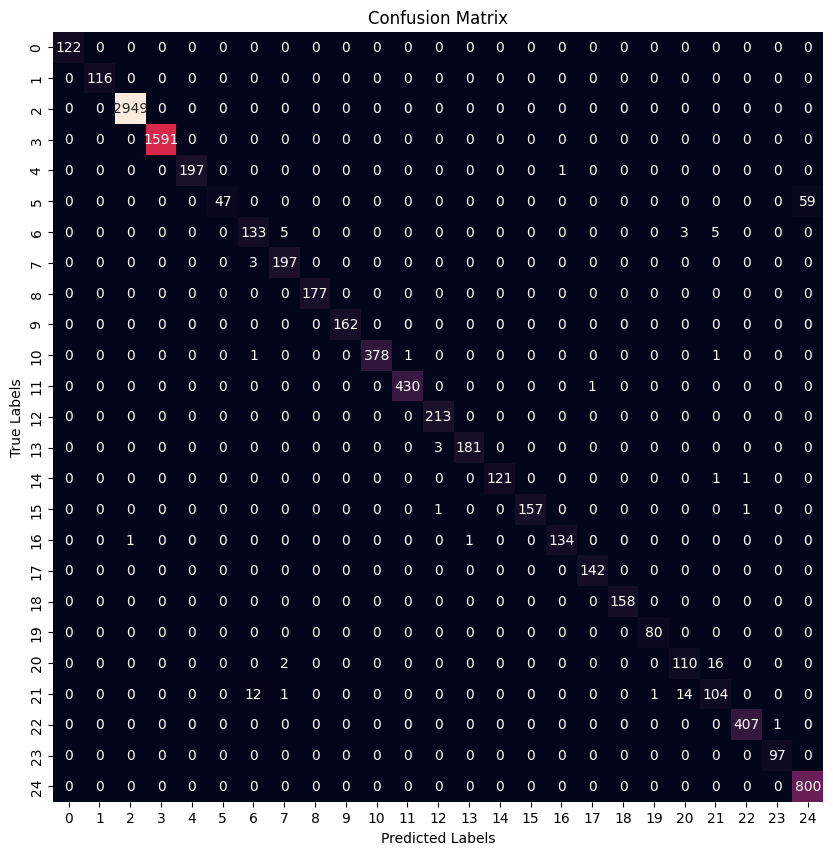
| Precision | Recall | F1 Score | |
|---|---|---|---|
| 0 | 1.000000 | 1.000000 | 1.000000 |
| 1 | 1.000000 | 1.000000 | 1.000000 |
| 2 | 0.999661 | 1.000000 | 0.999830 |
| 3 | 1.000000 | 1.000000 | 1.000000 |
| 4 | 1.000000 | 0.994949 | 0.997468 |
| 5 | 1.000000 | 0.443396 | 0.614379 |
| 6 | 0.892617 | 0.910959 | 0.901695 |
| 7 | 0.960976 | 0.985000 | 0.972840 |
| 8 | 1.000000 | 1.000000 | 1.000000 |
| 9 | 1.000000 | 1.000000 | 1.000000 |
| 10 | 1.000000 | 0.992126 | 0.996047 |
| 11 | 0.997680 | 0.997680 | 0.997680 |
| 12 | 0.981567 | 1.000000 | 0.990698 |
| 13 | 0.994505 | 0.983696 | 0.989071 |
| 14 | 1.000000 | 0.983740 | 0.991803 |
| 15 | 1.000000 | 0.987421 | 0.993671 |
| 16 | 0.992593 | 0.985294 | 0.988930 |
| 17 | 0.993007 | 1.000000 | 0.996491 |
| 18 | 1.000000 | 1.000000 | 1.000000 |
| 19 | 0.987654 | 1.000000 | 0.993789 |
| 20 | 0.866142 | 0.859375 | 0.862745 |
| 21 | 0.818898 | 0.787879 | 0.803089 |
| 22 | 0.995110 | 0.997549 | 0.996328 |
| 23 | 0.989796 | 1.000000 | 0.994872 |
| 24 | 0.931315 | 1.000000 | 0.964436 |
As it can be seen both architectures results in very similar f1 scores where some families are classified better than others.
Best Model on Bigger Dataset
Once we found the best model it was applied to the larger dataset. Naturally the results are not nearly as good as the MalImg dataset given that for that particular dataset there are only 25 families and there are fewer images. However, the model still yielded reasonable results. The 10-fold cross validation of the best model on Akash’s dataset yielded the following resutls:
| Folds | Accuracy | MAE |
|---|---|---|
| Fold 1 | 89% | 18.68 |
| Fold 2 | 89% | 18.72 |
| Fold 3 | 89% | 19.08 |
| Fold 4 | 90% | 17.50 |
| Fold 5 | 88% | 20.00 |
| Fold 6 | 89% | 18.14 |
| Fold 7 | 89% | 19.20 |
| Fold 8 | 89% | 18.51 |
| Fold 9 | 89% | 18.68 |
| Fold 10 | 89% | 19.45 |
| Average | 89.47% | 18.80 |
| Standard Deviation | 0.447% | 0.657 |
References
- Gibert, Daniel & Mateu, Carles & Planes, Jordi & Vicens, Ramon. (2019). Using convolutional neural networks for classification of malware represented as images. Journal of Computer Virology and Hacking Techniques. 15. 10.1007/s11416-018-0323-0.